mBuild¶
A hierarchical, component based molecule builder
With just a few lines of mBuild code, you can assemble reusable components into complex molecular systems for molecular simulations.
- mBuild is designed to minimize or even eliminate the need to explicitly translate and orient components when building systems: you simply tell it to connect two pieces!
- mBuild keeps track of the system’s topology so you don’t have to worry about manually defining bonds when constructing chemically bonded structures from smaller components.
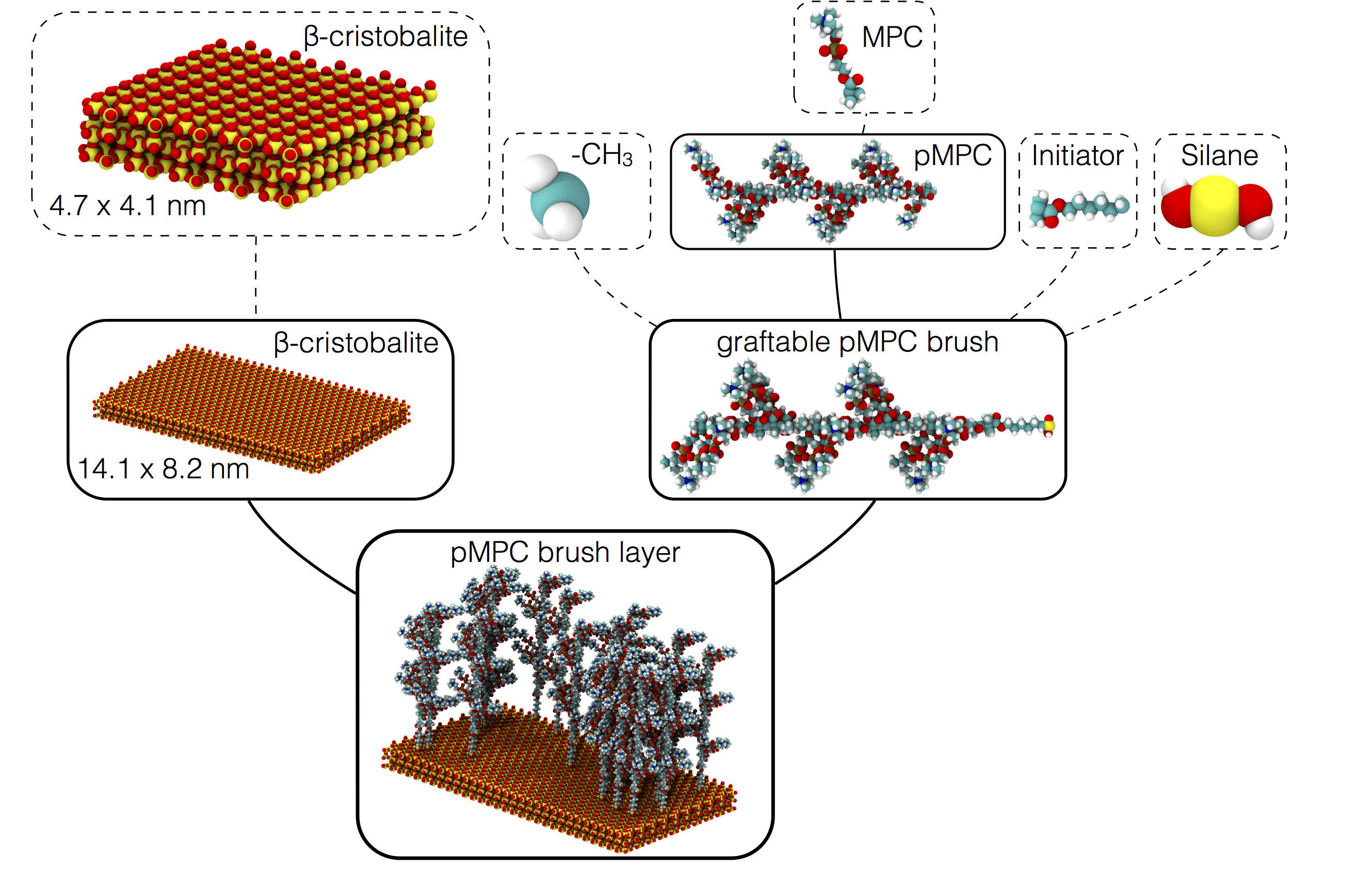
Example system¶
Components in dashed boxes are drawn by hand using, e.g., Avogadro or generated elsewhere. Each component is wrapped as a simple python class with user defined attachment sites, or ports. That’s the hard part! Now mBuild can do the rest. Each component further down the hierarchy is, again, a simple python class that describes which piece should connect to which piece.
Ultimately, complex systems structures can be created with just a line or two of code. Additionally, this approach seamlessly exposes tunable parameters within the hierarchy so you can actually create whole families of structures simply by adjusting a variable:
pattern = Random2DPattern(20) # A random arrangement of 20 pieces on a 2D surface.
brush_layer = BrushLayer(chain_lenth=20, pattern=pattern, tile_x=3, tile_y=2)

Various sub-portions of this library may be independently distributed under different licenses. See those files for their specific terms.
- Installation
- Using mBuild with Docker
- Tutorials
- Data Structures
- Coordinate transformations
- Recipes
- Citing mBuild
- Older Documentation